This week we hear from volunteer Stephen Chandler, who has been supporting The Microverse project by using computer software to identify the taxonomic groupings of the DNA sequences revealed in the sequencing machine.
Due to the size of microorganisms, we have until recent years relied on microscopes to identify different species. The advancement of scientific technologies however has made it possible for scientists to extract DNA from microorganisms, amplify that DNA into large quantities and then put the samples into a sequencing machine to reveal the genetic sequences. In The Microverse project, my role begins when the sequencer has finished processing the samples.
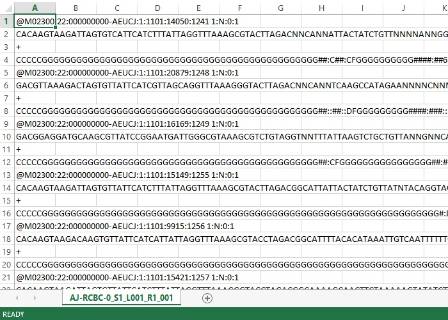
A raw data file from the MiSeq machine.
When the gene sequencer has finished decoding the PCR products it creates a file much like a typical excel file. The main difference is that this file can be incredibly large as it contains millions of DNA sequences belonging to hundreds if not thousands of species. This requires a powerful computer to run the analysis to identify what is in the sample.
At the Museum we use a number of servers with huge memory capacities and processing capabilities. To give an idea of the power these machines have compared to an everyday computer; a server at the Museum has at least 1.5TB (Terabytes) of RAM, that’s 300 times more processing power than your average computer, which has 4-6GB (Gigabytes) of RAM.
In order to use this computing power, the server needs to have a program designed to analyse and identify the DNA sequences, using a reference database of DNA for that group of organisms. To do this I use a program called QIIME (Quantative Insights Into Microbial Ecology).
The QIIME terminal, where the computer code is inputed to process the sequences.
The process of turning a raw sequence file listing all the DNA sequences, hot from the gene sequencer, into something that can be used to create graphs is not an easy task, especially when you have hundreds of thousands of sequences, as for the Microverse project.
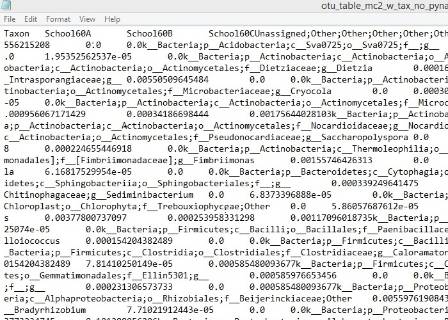
The first step is to remove low quality sequences that have errors. Then the sequences within a sample are grouped together into Operational Taxonomic Units (OTUs), according to their similarity. Sequences that are at least 97% similar to each other are grouped into one undefined OTU. The OTUs that are found are then compared to a reference database containing hundreds of thousands of specific species, and other taxonomic groupings, to identify which type of organisms they are.
A nearly completed file. All the sequences have been identified, but now need to be put into an order.
Some of the bacteria that we find are common and you can find them living on most surfaces in our home or garden, but others are incredibly rare and have evolved to survive in the most competitive and extreme environments. And all this microscopic life and diversity can all be found living just outside the front door. Although in the Microverse project no sample or result seems to be quite the same, which makes this a very exciting project.
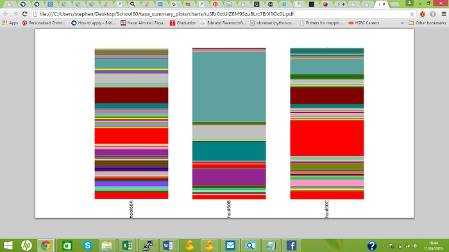
Three coloumn graphs representing the relative abundance of different microorganisms identified in three different samples.
Stephen Chandler
Stephen Chandler obtained a degree in marine biology at Portsmouth University and then went on to complete his masters at Imperial College London in ecology, conservation, and evolution in 2014. Stephen’s ambition is to study for a PhD and he is particularly interested in studying microorganisms in marine environments.
Stephen taking samples from the pocket roof of St Paul's Cathedral.
And now a brief word from Dr. Anne Jungblut, on careers in genomic science:
More and more research in biology, ecology and medicine is based on DNA and genome sequencing. The research relies on specialist software and programming in order to be able to analyse data sets as big as the Microverse sequence data, with future genomics projects likely to be much much bigger than our current project.
Along with specialist software the field will also need more and more different types of experts working on DNA projects to tackle future challenges in science, ranging from people interested in going outside to collect field data, molecular biologists that know how to do laboratory work to extract high quality DNA and run sequencing machines, to people that love concentrating on data analysis by applying specialist software, writing programming scripts or even develop new bioinformatics programs.
Anne Jungblut