Hello! I'm Filipa, the laboratory assistant in the Microverse project. My role is to prepare all the samples that arrive from schools and community groups for DNA sequencing.
Each group collected 10 samples from three different locations, which they labelled A, B and C. I select one sample from each location and I set up my lab bench with everything I need, including micropippetes, tubes and the reagents necessary for DNA extraction.
Filipa's workbench, ready to extract DNA from the samples.
Then I label all the tubes I'm going to use with the respective sample code, so that none of the samples gets mixed up, otherwise that would lead to misleading results. Then I extract the cotton wool, where all microorganisms are, from the wooden stick with the help of a pair of forceps and I use the reagents - following a specific protocol - to extract the DNA from the microorganisms. Finally I get a tube with DNA in it!
DNA extracted from the microorganisms.
We then use this DNA to carry out a PCR (Polimerase Chain Reaction) - a process through which we are able to amplify a specific DNA region, by producing millions of copies. We chose to sequence the gene for the 16S rRNA, which is regarded to be an excellent genetic marker for microbial community biodiversity studies due to it being an essential component of the protein synthesis machinery. That will enable us to identify which microorganisms are present in the sample. We amplify each sample three times (with different DNA concentrations), plus a negative control (with no DNA) to make sure that there isn't any contamination in the reaction.
This is the machine to visualise PCR products on an agarose gel using electrophoresis.
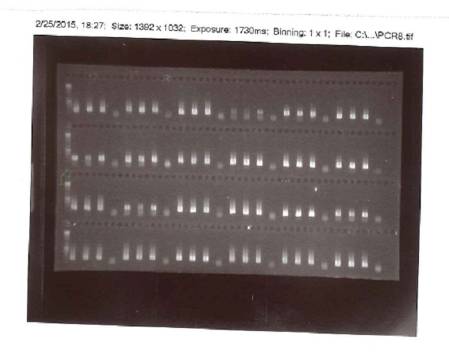
Then we run the PCR products on an agarose gel to see whether we have amplified the right size fragment - we expect our gene (16S rRNA) to be a 300-350 base pair fragment, which we compare with the ladder on the left - and that the control sample does not show up at all. The result is something like this:
A photograph of the PCR products after gel electrophoresis.
Everything worked! For each sample we have three bright bands in the position for 300-350 base pairs and a blank one, where we put the control.
Lets imagine that this was not the case and some things hadn't worked so well. For instance, if we didn't get a bright band from our samples it would mean that the DNA fragment wasn't amplified. In this case it would mean that an error occurred during the PCR set up and as a result we would need to repeat it.
It could also happen that we found a band in one of our negative controls, this would reveal a contamination in the PCR reagents, which are not supposed to have any DNA. To solve this, we would need to start again with brand new reagents (and be more careful!).
In the control sample, and in some of the samples with DNA, we see a short faint fragment, this is a by-product of the reaction called a primer-dimer. To remove this we do a PCR clean-up. When that is done the samples are almost ready to be sent for sequencing, and soon after we will find out what microorganisms inhabit the surfaces you've been swabbing!
Filipa Leao Sampaio, laboratory assistant.
Filipa is a laboratory assistant at the Museum, she began her career with an undergraduate degree in Biology and then a masters in Biodiversity, Genetics and Evolution in the University of Porto, in Portugal. For her dissertation she worked on a project where she studied phylogenetic relationships and patterns of genetic diversity in reptiles from the Mediterranean Basin.
Since September 2013 she has been working at the Museum carrying out molecular lab work on different projects - snake vision evolution, Antarctic soil microbial diversity and UK urban microbial diversity. Later this year she starts a PhD in London where she will receive training in different areas of environmental sciences.
Jade Lauren