Today one of our Microverse citizen science project participants, Robert Milne, presents his own interpretation of the results of the microbial samples collected from Mid Kent College in Gillingham where he is a student:
The results:
Despite our best efforts, the samples we obtained for the Microverse project were taken in different weather conditions, at slightly different times, in slightly different areas of the building, and all three samples were taken from walls facing different directions. The materials of the surfaces we sampled were brick, glass and metal.
Mid Kent College building, swabbed by The Microverse participants.
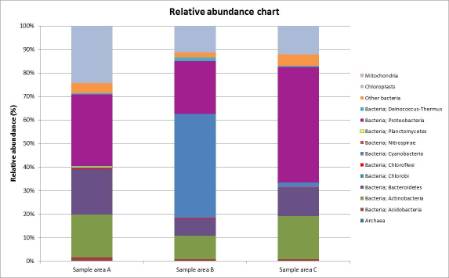
From the results below, it can be seen that all three surfaces have about the same number of OTUs, (Operational Taxonomic Units, a phrase to indicate taxonomic groupings in microorganisms), but this does not mean that each surface has the same number of individual microorganisms. The number of genetic sequences varies greatly.
Sample Area A (brick) | Sample Area B (glass) | Sample Area C (metal) | |
|---|---|---|---|
| Number of genetic sequences generated | 88,264 | 120,498 | 27,894 |
| Number of OTUs | 2,198 | 2,107 | 1,960 |
| % of sequences that were from Archaea | 0.02% | 0.00% | 0.00% |
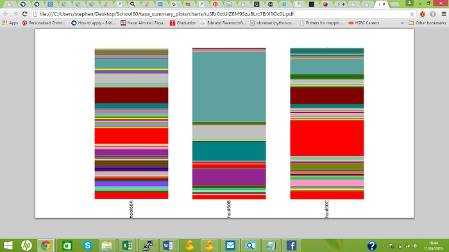
| % of sequences that were from Bacteria | 75.62% | 88.76% | 87.75% |
| % of sequences that were from Eukaryotes | 24.36% | 11.24% | 12.19% |
Table 1: Results from samples of microorganisms swabbed from brick, glass and metal, at Mid Kent College, Gillingham, (% rounded to 2 decimal places).
The glass surface has generated the most genetic sequences while metal has generated the least. This could mean that the bacteria on the surface of the glass are more successful than the ones on the metal, for instance.
Sample Area A - Brick.
The image above shows the brick wall from which the first sample was taken. This wall had the most eukaryotic cells present, in which the majority of them contained chloroplasts (these are the organelles of plants that convert light energy into sugar).
This wall faces southwest and a wall facing south of any kind will always receive the most sunlight on it during the day, which could explain the increased chloroplast numbers compared to the other two surface areas we sampled. The fact that this wall was also close to a lot of grass could also play a part in these numbers.
Sample Area B - Glass.
The image above shows the second surface sampled, which was glass. This had the most genetic sequences found out of all three of the surfaces we swabbed. There were, however, less eukaryotic cells on the glass and metal surfaces than on the wall.
This could be because the smooth surface of the metal and the glass meant that less eukaryote cells could remain on the surfaces for prolonged periods. The eukaryotic cells (represented by the mitochondria and chloroplast sequences in the sample) could have originated from natural wildlife around the area, such as a snail's trail or some spider webbing.
Sample Area C - Metal.
Most of the eukaryote sequences found in all samples were chloroplasts, rather than mitochondria. This probably means the surfaces always have some form of sunlight on them, which is somewhat true since all the surfaces faced either west or east to some extent.
Figure 1: The relative abundance of bacterial phyla, archaea, mitochondria and chloroplasts in the three samples.
Possible uses:
One of the prime examples for undertaking this feat of exploring more of the microbiological world is the need to find better antibiotics; resistance to antibiotics is an increasing threat in the world of medicine. Antibiotic discovery can occur via the identification of bacteria that produce chemical substances that kill or inhibit the growth of other bacteria. Once identified the chemical substance can potentially be cultured and used as a treatment to kill off bacterial infections.
Exploring the countless surfaces outside in the world is a treasure trove of information that could lead to the discoveries of new bacteria that can be used effectively as a source for an antibiotic.
However, it can also be considered that a new resilient bacteria could be discovered that can survive without much water for a long time, which may, just maybe, hold a specific DNA sequence to help relieve the effects of hunger and thirst in patients that must undergo a fast before an operation (such as colon screening). It can open up a number of new doors to the world of medicine, and with a huge percent of areas still not investigated, it could only be a matter of time before huge changes are discovered.
Robert Milne
Thank you Robert! Robert Milne is a student of Mid Kent College, who has just finished his second year of an Applied Science Level 3 course. He has a keen interest in biochemistry and genetics and hopes to enrol this Autumn on an Undergraduate degree in Chemistry at the University of Greenwich. To find out more about the Museum's citizen science projects, see our website.